|
|
|
|
-
pathway class
-
N-glycan
- <a href="gg142">ALG1</a>
- <a href="gg143">ALG2</a>
- <a href="gg144">ALG3</a>
- <a href="gg145">ALG5</a>
- <a href="gg146">ALG6</a>
- <a href="gg147">ALG8</a>
- <a href="gg148">ALG9</a>
- <a href="gg149">ALG10</a>
- <a href="gg201">ALG11</a>
- <a href="gg151">ALG12</a>
- <a href="gg205">ALG13</a>
- <a href="gg202">ALG14</a>
- <a href="gg077">B4GALT1</a>
- <a href="gg078">B4GALT2</a>
- <a href="gg079">B4GALT3</a>
- <a href="gg122">B3GNT8</a>
- <a href="gg105">B4GALNT2</a>
- <a href="gg141">DPAGT1</a>
- <a href="gg140">DPM1</a>
- <a href="gg203">DPM2</a>
- <a href="gg204">DPM3</a>
- <a href="gg009">FUT8</a>
- <a href="gg010">FUT10</a>
- <a href="gg125">MGAT1</a>
- <a href="gg126">MGAT2</a>
- <a href="gg127">MGAT3</a>
- <a href="gg128">MGAT4A</a>
- <a href="gg129">MGAT4B</a>
- <a href="gg130">MGAT5</a>
- <a href="gg131">MGAT5B</a>
- <a href="gg054">ST6GAL1</a>
- <a href="gg055">ST6GAL2</a>
- <a href="gg063">ST8SIA2</a>
- <a href="gg064">ST8SIA3</a>
- <a href="gg065">ST8SIA4</a>
- <a href="gg045">CHST8</a>
- <a href="gg046">CHST9</a>
- <a href="gg152">UGGT1 (UGCGL1)</a>
- <a href="gg153">UGGT2 (UGCGL2)</a>
-
O-glycan, mucin-type
- <a href="gg120">B3GNT6</a>
- <a href="gg081">B4GALT5</a>
- <a href="gg124">A4GNT</a>
- <a href="gg105">B4GALNT2</a>
- <a href="gg075">C1GALT1</a>
- <a href="gg076">C1GALT1C1</a>
- <a href="gg087">GALNT1</a>
- <a href="gg088">GALNT2</a>
- <a href="gg089">GALNT3</a>
- <a href="gg090">GALNT4</a>
- <a href="gg091">GALNT5</a>
- <a href="gg092">GALNT6</a>
- <a href="gg093">GALNT7</a>
- <a href="gg094">GALNT8</a>
- <a href="gg095">GALNT9</a>
- <a href="gg096">GALNT10</a>
- <a href="gg097">GALNT11</a>
- <a href="gg098">GALNT12</a>
- <a href="gg099">GALNT13</a>
- <a href="gg100">GALNT14</a>
- <a href="gg101">GALNT15</a>
- <a href="gg212">GALNT16</a>
- <a href="gg211">GALNT17</a>
- <a href="gg183">GALNT18</a>
- <a href="gg207">GALNT19</a>
- <a href="gg181">GALNT20</a>
- <a href="gg109">GCNT1</a>
- <a href="gg108">GCNT3</a>
- <a href="gg110">GCNT4</a>
- <a href="gg048">ST3GAL1</a>
- <a href="gg049">ST3GAL2</a>
- <a href="gg056">ST6GALNAC1</a>
- <a href="gg057">ST6GALNAC2</a>
- <a href="gg065">ST8SIA4</a>
- <a href="gg067">ST8SIA6</a>
-
O-glycan, not mucin-type
- <a href="gg184">B3GLCT</a>
- <a href="gg157">B3GAT1</a>
- <a href="gg156">B3GAT2</a>
- <a href="gg077">B4GALT1</a>
- <a href="gg078">B4GALT2</a>
- <a href="gg079">B4GALT3</a>
- <a href="gg080">B4GALT4</a>
- <a href="gg103">B3GALNT2</a>
- <a href="gg035">CHST10</a>
- <a href="gg210">EOGT</a>
- <a href="gg187">FCMD</a>
- <a href="gg188">FKRP</a>
- <a href="gg004">FUT4</a>
- <a href="gg007">FUT7</a>
- <a href="gg008">FUT9</a>
- <a href="gg172">GYLTL1B</a>
- <a href="gg185">GXYLT1</a>
- <a href="gg186">GXYLT2</a>
- <a href="gg171">LARGE</a>
- <a href="gg133">LFNG</a>
- <a href="gg131">MGAT5B</a>
- <a href="gg132">MFNG</a>
- <a href="gg114">OGT</a>
- <a href="gg138">POMT1</a>
- <a href="gg139">POMT2</a>
- <a href="gg012">POFUT1</a>
- <a href="gg013">POFUT2</a>
- <a href="gg208">POGLUT1</a>
- <a href="gg123">POMGNT1</a>
- <a href="gg134">RFNG</a>
- <a href="gg052">ST3GAL5</a>
- <a href="gg054">ST6GAL1</a>
- <a href="gg055">ST6GAL2</a>
- <a href="gg209">XXYLT1</a>
- <a href="gg133">LFNG</a>
- <a href="gg132">MFNG</a>
- <a href="gg134">RFNG</a>
- <a href="gg235">POMGNT2</a>
- <a href="gg236">POMK</a>
-
Glycosaminoglycan, chondroitin sulfate
- <a href="gg074">B3GALT6</a>
- <a href="gg155">B3GAT3</a>
- <a href="gg083">B4GALT7</a>
- <a href="gg167">CHPF</a>
- <a href="gg164">CSGlcA-T</a>
- <a href="gg025">CHST3</a>
- <a href="gg026">CHST7</a>
- <a href="gg036">CHST11</a>
- <a href="gg037">CHST12</a>
- <a href="gg038">CHST13</a>
- <a href="gg040">D4ST1</a>
- <a href="gg047">GALNAC4S-6ST</a>
- <a href="gg163">CHSY1</a>
- <a href="gg168">CSS3</a>
- <a href="gg165">CSGALNACT1</a>
- <a href="gg166">CSGALNACT2</a>
- <a href="gg039">UST</a>
- <a href="gg169">XYLT1</a>
- <a href="gg170">XYLT2</a>
-
Glycosaminoglycan, heparan sulfate
- <a href="gg155">B3GAT3</a>
- <a href="gg074">B3GALT6</a>
- <a href="gg083">B4GALT7</a>
- <a href="gg158">EXT1</a>
- <a href="gg159">EXT2</a>
- <a href="gg160">EXTL1</a>
- <a href="gg161">EXTL2</a>
- <a href="gg162">EXTL3</a>
- <a href="gg024">HS2ST1</a>
- <a href="gg014">HS3ST1</a>
- <a href="gg015">HS3ST2</a>
- <a href="gg016">HS3ST3A1</a>
- <a href="gg017">HS3ST3B1</a>
- <a href="gg018">HS3ST4</a>
- <a href="gg019">HS3ST5</a>
- <a href="gg031">HS6ST1</a>
- <a href="gg032">HS6ST2</a>
- <a href="gg033">HS6ST3</a>
- <a href="gg020">NDST1</a>
- <a href="gg021">NDST2</a>
- <a href="gg022">NDST3</a>
- <a href="gg023">NDST4</a>
- <a href="gg169">XYLT1</a>
- <a href="gg170">XYLT2</a>
-
Glycosaminoglycan, keratan sulfate
- <a href="gg115">B4GAT1</a>
- <a href="gg116">B3GNT2</a>
- <a href="gg121">B3GNT7</a>
- <a href="gg077">B4GALT1</a>
- <a href="gg078">B4GALT2</a>
- <a href="gg079">B4GALT3</a>
- <a href="gg080">B4GALT4</a>
- <a href="gg027">CHST1</a>
- <a href="gg028">CHST2</a>
- <a href="gg034">CHST4</a>
- <a href="gg029">CHST5</a>
- <a href="gg030">CHST6</a>
- <a href="gg009">FUT8</a>
- <a href="gg048">ST3GAL1</a>
- <a href="gg049">ST3GAL2</a>
- <a href="gg050">ST3GAL3</a>
-
GPI-anchor
- <a href="gg203">DPM2</a>
- <a href="gg231">GAA1</a>
- <a href="gg215">PIGA</a>
- <a href="gg225">PIGB</a>
- <a href="gg214">PIGC</a>
- <a href="gg228">PIGF</a>
- <a href="gg229">PIGG</a>
- <a href="gg216">PIGH</a>
- <a href="gg230">PIGK</a>
- <a href="gg219">PIGL</a>
- <a href="gg221">PIGM</a>
- <a href="gg224">PIGN</a>
- <a href="gg227">PIGO</a>
- <a href="gg217">PIGP</a>
- <a href="gg213">PIGQ</a>
- <a href="gg232">PIGS</a>
- <a href="gg233">PIGT</a>
- <a href="gg234">PIGU</a>
- <a href="gg223">PIGV</a>
- <a href="gg220">PIGW</a>
- <a href="gg222">PIGX</a>
- <a href="gg218">PIGY</a>
- <a href="gg226">PIGZ</a>
-
glycolipid, lacto/neolacto series
- <a href="gg200">A(ABO)</a>
- <a href="gg085">B(ABO)</a>
- <a href="gg070">B3GALT1</a>
- <a href="gg071">B3GALT2</a>
- <a href="gg073">B3GALT5</a>
- <a href="gg115">B4GAT1</a>
- <a href="gg116">B3GNT2</a>
- <a href="gg117">B3GNT3</a>
- <a href="gg118">B3GNT4</a>
- <a href="gg119">B3GNT5</a>
- <a href="gg077">B4GALT1</a>
- <a href="gg078">B4GALT2</a>
- <a href="gg079">B4GALT3</a>
- <a href="gg080">B4GALT4</a>
- <a href="gg001">FUT1</a>
- <a href="gg002">FUT2</a>
- <a href="gg003">FUT3</a>
- <a href="gg004">FUT4</a>
- <a href="gg005">FUT5</a>
- <a href="gg006">FUT6</a>
- <a href="gg007">FUT7</a>
- <a href="gg008">FUT9</a>
- <a href="gg111">GCNT2</a>
- <a href="gg050">ST3GAL3</a>
- <a href="gg051">ST3GAL4</a>
- <a href="gg053">ST3GAL6</a>
- <a href="gg062">ST8SIA1</a>
-
glycolipid, ganglio series
- <a href="gg072">B3GALT4</a>
- <a href="gg104">B4GALNT1</a>
- <a href="gg105">B4GALNT2</a>
- <a href="gg048">ST3GAL1</a>
- <a href="gg049">ST3GAL2</a>
- <a href="gg052">ST3GAL5</a>
- <a href="gg058">ST6GALNAC3</a>
- <a href="gg059">ST6GALNAC4</a>
- <a href="gg060">ST6GALNAC5</a>
- <a href="gg061">ST6GALNAC6</a>
- <a href="gg062">ST8SIA1</a>
- <a href="gg064">ST8SIA3</a>
- <a href="gg066">ST8SIA5</a>
-
glycolipid, globo series
- <a href="gg068">A4GALT</a>
- <a href="gg073">B3GALT5</a>
- <a href="gg102">B3GALNT1</a>
- <a href="gg001">FUT1</a>
- <a href="gg002">FUT2</a>
- <a href="gg008">FUT9</a>
- <a href="gg086">GBGT1</a>
- <a href="gg048">ST3GAL1</a>
- <a href="gg049">ST3GAL2</a>
- <a href="gg062">ST8SIA1</a>
-
Hyaluronic acid
- <a href="gg135">HAS1</a>
- <a href="gg136">HAS2</a>
- <a href="gg137">HAS3</a>
-
sphingolipid
- <a href="gg082">B4GALT6</a>
- <a href="gg041">GAL3ST1</a>
- <a href="gg084">CGT</a>
- <a href="gg154">UGCG</a>
-
SLC/transporter
- <a href="gg178">SLC35A1</a>
- <a href="gg176">SLC35A2</a>
- <a href="gg174">SLC35A3</a>
- <a href="gg175">SLC35B1</a>
- <a href="gg179">SLC35B2</a>
- <a href="gg192">SLC35B3</a>
- <a href="gg206">SLC35B4</a>
- <a href="gg173">SLC35C1</a>
- <a href="gg177">SLC35D1</a>
- <a href="gg180">SLC35D2</a>
-
terminal modification
- <a href="gg106">B4GALNT3</a>
- <a href="gg107">B4GALNT4</a>
- <a href="gg042">GAL3ST2</a>
- <a href="gg043">GAL3ST3</a>
- <a href="gg044">GAL3ST4</a>
-
unassign
- <a href="gg011">FUT11</a>
-
additional
- SFT2
- SFT3
- COLGALT2
- MGAT4C
- LHL2
- L5
- HS3ST6
- COLGALT1
- L4_nuc
- B3GNT9
- SFT9
- A3GALT2
- UGT3A1
- GCNT7
- ALG10B
- ALG1L
- ALG1L2
- SEC1P
- C5
- TMEM5
- ART1
-
Enzymatic Activity
-
ALG
-
Doner Synthesis
-
Dol-P-Man Synthesis
- <a href="gg140">DPM1</a>
- <a href="gg203">DPM2</a>
- <a href="gg204">DPM3</a>
-
Dol-P-Glc Synthesis
-
Cytoplasmic ALG(GnT)
- <a href="gg202">ALG14</a>
- <a href="gg141">DPAGT1</a>
- <a href="gg205">ALG13</a>
-
Cytoplasmic ALG(ManT)
- <a href="gg142">ALG1</a>
- <a href="gg201">ALG11</a>
- <a href="gg143">ALG2</a>
-
ER Lumen ALG(ManT)
- <a href="gg151">ALG12</a>
- <a href="gg144">ALG3</a>
- <a href="gg148">ALG9</a>
-
ER Lumen ALG(GlcT)
- <a href="gg149">ALG10</a>
- <a href="gg146">ALG6</a>
- <a href="gg147">ALG8</a>
-
Glucosyltransferases
- <a href="gg154">UGCG</a>
- <a href="gg152">UGGT1 (UGCGL1)</a>
- <a href="gg153">UGGT2 (UGCGL2)</a>
-
Xylosyltransferases
- <a href="gg169">XYLT1</a>
- <a href="gg170">XYLT2</a>
- <a href="gg185">GXYLT1</a>
- <a href="gg186">GXYLT2</a>
- <a href="gg209">XXYLT1</a>
- <a href="gg171">LARGE</a>
- <a href="gg172">GYLTL1B</a>
-
Fucosyltransferases
- <a href="gg001">FUT1</a>
- <a href="gg002">FUT2</a>
- <a href="gg003">FUT3</a>
- <a href="gg004">FUT4</a>
- <a href="gg005">FUT5</a>
- <a href="gg006">FUT6</a>
- <a href="gg007">FUT7</a>
- <a href="gg009">FUT8</a>
- <a href="gg008">FUT9</a>
-
O-Fucosyltansferases
- <a href="gg012">POFUT1</a>
- <a href="gg013">POFUT2</a>
-
Fucosyltransferases (Unknown Activities)
- <a href="gg010">FUT10</a>
- <a href="gg011">FUT11</a>
-
Mannosyltransferases
- <a href="gg138">POMT1</a>
- <a href="gg139">POMT2</a>
- <a href="gg221">PIGM</a>
- <a href="gg222">PIGX</a>
- <a href="gg223">PIGV</a>
- <a href="gg225">PIGB</a>
- <a href="gg226">PIGZ</a>
-
Galactosyltransferases
-
Ceramide
-
alpha
- <a href="gg068">A4GALT</a>
- <a href="gg085">B(ABO)</a>
-
beta 1,3
- <a href="gg070">B3GALT1</a>
- <a href="gg071">B3GALT2</a>
- <a href="gg072">B3GALT4</a>
- <a href="gg073">B3GALT5</a>
- <a href="gg074">B3GALT6</a>
- <a href="gg075">C1GALT1</a>
-
beta 1,4
- <a href="gg077">B4GALT1</a>
- <a href="gg078">B4GALT2</a>
- <a href="gg079">B4GALT3</a>
- <a href="gg080">B4GALT4</a>
- <a href="gg081">B4GALT5</a>
- <a href="gg082">B4GALT6</a>
- <a href="gg083">B4GALT7</a>
-
N-Acetylgalactosaminyltransferases
-
beta 1,3
- <a href="gg102">B3GALNT1</a>
- <a href="gg103">B3GALNT2</a>
-
beta 1,4
- <a href="gg104">B4GALNT1</a>
- <a href="gg105">B4GALNT2</a>
- <a href="gg106">B4GALNT3</a>
- <a href="gg107">B4GALNT4</a>
-
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases
- <a href="gg087">GALNT1</a>
- <a href="gg088">GALNT2</a>
- <a href="gg089">GALNT3</a>
- <a href="gg090">GALNT4</a>
- <a href="gg091">GALNT5</a>
- <a href="gg092">GALNT6</a>
- <a href="gg093">GALNT7</a>
- <a href="gg094">GALNT8</a>
- <a href="gg095">GALNT9</a>
- <a href="gg096">GALNT10</a>
- <a href="gg097">GALNT11</a>
- <a href="gg098">GALNT12</a>
- <a href="gg099">GALNT13</a>
- <a href="gg100">GALNT14</a>
- <a href="gg101">GALNT15</a>
- <a href="gg212">GALNT16</a>
- <a href="gg211">GALNT17</a>
- <a href="gg183">GALNT18</a>
- <a href="gg207">GALNT19</a>
- <a href="gg181">GALNT20</a>
-
alpha 1,3
- <a href="gg200">A(ABO)</a>
- <a href="gg086">GBGT1</a>
-
N-Acetylglucosaminyltransferases
-
alpha 1,4
- <a href="gg124">A4GNT</a>
-
alpha 1,6
- <a href="gg213">PIGQ</a>
- <a href="gg214">PIGC</a>
- <a href="gg215">PIGA</a>
- <a href="gg216">PIGH</a>
- <a href="gg217">PIGP</a>
- <a href="gg218">PIGY</a>
- <a href="gg219">PIGL</a>
- <a href="gg220">PIGW</a>
-
beta 1,3
- <a href="gg115">B4GAT1</a>
- <a href="gg116">B3GNT2</a>
- <a href="gg117">B3GNT3</a>
- <a href="gg118">B3GNT4</a>
- <a href="gg119">B3GNT5</a>
- <a href="gg120">B3GNT6</a>
- <a href="gg121">B3GNT7</a>
- <a href="gg122">B3GNT8</a>
-
beta 1,6 (core2)
- <a href="gg109">GCNT1</a>
- <a href="gg108">GCNT3</a>
- <a href="gg110">GCNT4</a>
-
blood I antigen
- <a href="gg111">GCNT2</a>
-
MGAT family
- <a href="gg125">MGAT1</a>
- <a href="gg126">MGAT2</a>
- <a href="gg127">MGAT3</a>
- <a href="gg128">MGAT4A</a>
- <a href="gg129">MGAT4B</a>
- <a href="gg130">MGAT5</a>
- <a href="gg131">MGAT5B</a>
-
O-linked
- <a href="gg114">OGT</a>
- <a href="gg210">EOGT</a>
-
Protein O-linked fucose b1,3-N-acetylglucosaminyltransferases
- <a href="gg132">MFNG</a>
- <a href="gg133">LFNG</a>
- <a href="gg134">RFNG</a>
-
Protein O-linked mannose b1,2-N-acetylglucosaminyltransferases
- <a href="gg123">POMGNT1</a>
-
Protein O-linked mannose b1,4-N-acetylglucosaminyltransferases
- <a href="gg235">POMGNT2</a>
-
Glucosyltransferase
- <a href="gg184">B3GLCT</a>
- <a href="gg208">POGLUT1</a>
- <a href="gg171">LARGE</a>
- <a href="gg172">GYLTL1B</a>
-
Sialyltransferases
-
beta-Gal alpha 2,3-sialyltransferases
- <a href="gg048">ST3GAL1</a>
- <a href="gg049">ST3GAL2</a>
- <a href="gg050">ST3GAL3</a>
- <a href="gg051">ST3GAL4</a>
- <a href="gg052">ST3GAL5</a>
- <a href="gg053">ST3GAL6</a>
-
beta-Gal alpha 2,6-sialyltransferases
- <a href="gg054">ST6GAL1</a>
- <a href="gg055">ST6GAL2</a>
-
beta-GalNAc alpha 2,6-sialyltransferases
- <a href="gg056">ST6GALNAC1</a>
- <a href="gg057">ST6GALNAC2</a>
- <a href="gg058">ST6GALNAC3</a>
- <a href="gg059">ST6GALNAC4</a>
- <a href="gg060">ST6GALNAC5</a>
- <a href="gg061">ST6GALNAC6</a>
-
alpha NeuAc alpha 2,8-sialyltransferases
- <a href="gg062">ST8SIA1</a>
- <a href="gg063">ST8SIA2</a>
- <a href="gg064">ST8SIA3</a>
- <a href="gg065">ST8SIA4</a>
- <a href="gg066">ST8SIA5</a>
- <a href="gg067">ST8SIA6</a>
-
GAG
-
chondroitin
-
N-Acetylgalactosaminyltransferases
- <a href="gg165">CSGALNACT1</a>
- <a href="gg166">CSGALNACT2</a>
-
N-Acetylgalactosaminyltransferases Glucuronyltransferases GalNAc/GlcA
- <a href="gg167">CHPF</a>
- <a href="gg163">CHSY1</a>
- <a href="gg168">CSS3</a>
-
Glucuronyltransferases
- <a href="gg164">CSGlcA-T</a>
-
heparan sulfate
-
Glucuronyltransferases N-Acetylglucosaminyltransferases GlcA/GlcNAc
- <a href="gg158">EXT1</a>
- <a href="gg159">EXT2</a>
-
N-Acetylglucosaminyltransferases
- <a href="gg160">EXTL1</a>
- <a href="gg162">EXTL3</a>
- <a href="gg161">EXTL2</a>
-
Hyaluronan synthase
-
Glucuronyltransferases N-Acetylglucosaminyltransferases GAG GlcA/GlcNAc
- <a href="gg135">HAS1</a>
- <a href="gg136">HAS2</a>
- <a href="gg137">HAS3</a>
-
Glucuronyltransferases
- <a href="gg157">B3GAT1</a>
- <a href="gg156">B3GAT2</a>
- <a href="gg155">B3GAT3</a>
-
Sulfotransferases
-
HNK
- <a href="gg035">CHST10</a>
- <a href="gg034">CHST4</a>
- <a href="gg029">CHST5</a>
-
dermatan/chondroitin
-
Gal 2-O-sulfotransferase
-
Gal 3-O-sulfotransferase
- <a href="gg041">GAL3ST1</a>
- <a href="gg042">GAL3ST2</a>
- <a href="gg043">GAL3ST3</a>
- <a href="gg044">GAL3ST4</a>
-
GalNAc 4-O-sulfotransferase
- <a href="gg045">CHST8</a>
- <a href="gg046">CHST9</a>
- <a href="gg037">CHST12</a>
- <a href="gg038">CHST13</a>
- <a href="gg036">CHST11</a>
-
GalNAc4S 6-O-sulfotransferase
- <a href="gg047">GALNAC4S-6ST</a>
-
Keratan sulfate
-
Gal 6-O-sulfotransferase
- <a href="gg027">CHST1</a>
- <a href="gg025">CHST3</a>
- <a href="gg026">CHST7</a>
-
GlcNAc 6-O-sulfotransferase
- <a href="gg030">CHST6</a>
- <a href="gg028">CHST2</a>
-
dermatan sulfate
- <a href="gg040">D4ST1</a>
-
heparan sulfate
-
2-O-sulfotransferase
- <a href="gg024">HS2ST1</a>
-
3-O-sulfotransferase
- <a href="gg014">HS3ST1</a>
- <a href="gg015">HS3ST2</a>
- <a href="gg016">HS3ST3A1</a>
- <a href="gg017">HS3ST3B1</a>
- <a href="gg018">HS3ST4</a>
- <a href="gg019">HS3ST5</a>
-
6-O-sulfotransferase
- <a href="gg031">HS6ST1</a>
- <a href="gg032">HS6ST2</a>
- <a href="gg033">HS6ST3</a>
-
N-deacetylase/N-sulfotransferase
- <a href="gg020">NDST1</a>
- <a href="gg021">NDST2</a>
- <a href="gg022">NDST3</a>
- <a href="gg023">NDST4</a>
-
Nucleotide Sugar Transporters
-
CMP-sialic acid transporter
- <a href="gg178">SLC35A1</a>
-
UDP-galactose transporter
- <a href="gg176">SLC35A2</a>
- <a href="gg175">SLC35B1</a>
-
UDP-GlcNAc transporter
- <a href="gg174">SLC35A3</a>
-
PAPS transporter
- <a href="gg179">SLC35B2</a>
-
UDP-Xyl transporter and UDP-GlcNAc transporter
- <a href="gg206">SLC35B4</a>
-
GDP-fucose transporter
- <a href="gg173">SLC35C1</a>
-
UDP-GlcA transporter and UDP-GalNAc transporter
- <a href="gg177">SLC35D1</a>
-
UDP-GlcNAc transporter
- <a href="gg180">SLC35D2</a>
-
PAPS Transporter
- <a href="gg192">SLC35B3</a>
-
α-Dystroglycan Modification
- <a href="gg187">FCMD</a>
- <a href="gg188">FKRP</a>
-
GPI EtNP Transferase
- <a href="gg224">PIGN</a>
- <a href="gg227">PIGO</a>
- <a href="gg228">PIGF</a>
- <a href="gg229">PIGG</a>
-
GPI Transamidase
- <a href="gg230">PIGK</a>
- <a href="gg231">GAA1</a>
- <a href="gg232">PIGS</a>
- <a href="gg233">PIGT</a>
- <a href="gg234">PIGU</a>
-
Others
- <a href="gg236">POMK</a>
- <a href="gg076">C1GALT1C1</a>
-
EC class
-
2.4.1
- <a href="gg200">A(ABO)</a>
- <a href="gg068">A4GALT</a>
- <a href="gg142">ALG1</a>
- <a href="gg151">ALG12</a>
- <a href="gg143">ALG2</a>
- <a href="gg144">ALG3</a>
- <a href="gg145">ALG5</a>
- <a href="gg085">B(ABO)</a>
- <a href="gg102">B3GALNT1</a>
- <a href="gg072">B3GALT4</a>
- <a href="gg074">B3GALT6</a>
- <a href="gg184">B3GLCT</a>
- <a href="gg157">B3GAT1</a>
- <a href="gg156">B3GAT2</a>
- <a href="gg155">B3GAT3</a>
- <a href="gg116">B3GNT2</a>
- <a href="gg117">B3GNT3</a>
- <a href="gg118">B3GNT4</a>
- <a href="gg119">B3GNT5</a>
- <a href="gg120">B3GNT6</a>
- <a href="gg104">B4GALNT1</a>
- <a href="gg077">B4GALT1</a>
- <a href="gg078">B4GALT2</a>
- <a href="gg079">B4GALT3</a>
- <a href="gg080">B4GALT4</a>
- <a href="gg081">B4GALT5</a>
- <a href="gg082">B4GALT6</a>
- <a href="gg083">B4GALT7</a>
- <a href="gg075">C1GALT1</a>
- <a href="gg076">C1GALT1C1</a>
- <a href="gg084">CGT</a>
- <a href="gg165">CSGALNACT1</a>
- <a href="gg166">CSGALNACT2</a>
- <a href="gg164">CSGlcA-T</a>
- <a href="gg140">DPM1</a>
- <a href="gg203">DPM2</a>
- <a href="gg204">DPM3</a>
- <a href="gg158">EXT1</a>
- <a href="gg159">EXT2</a>
- <a href="gg160">EXTL1</a>
- <a href="gg161">EXTL2</a>
- <a href="gg162">EXTL3</a>
- <a href="gg001">FUT1</a>
- <a href="gg002">FUT2</a>
- <a href="gg003">FUT3</a>
- <a href="gg004">FUT4</a>
- <a href="gg005">FUT5</a>
- <a href="gg006">FUT6</a>
- <a href="gg007">FUT7</a>
- <a href="gg009">FUT8</a>
- <a href="gg008">FUT9</a>
- <a href="gg087">GALNT1</a>
- <a href="gg096">GALNT10</a>
- <a href="gg097">GALNT11</a>
- <a href="gg098">GALNT12</a>
- <a href="gg099">GALNT13</a>
- <a href="gg100">GALNT14</a>
- <a href="gg101">GALNT15</a>
- <a href="gg212">GALNT16</a>
- <a href="gg211">GALNT17</a>
- <a href="gg088">GALNT2</a>
- <a href="gg089">GALNT3</a>
- <a href="gg090">GALNT4</a>
- <a href="gg091">GALNT5</a>
- <a href="gg092">GALNT6</a>
- <a href="gg093">GALNT7</a>
- <a href="gg094">GALNT8</a>
- <a href="gg095">GALNT9</a>
- <a href="gg086">GBGT1</a>
- <a href="gg109">GCNT1</a>
- <a href="gg111">GCNT2</a>
- <a href="gg205">ALG13</a>
- <a href="gg135">HAS1</a>
- <a href="gg136">HAS2</a>
- <a href="gg137">HAS3</a>
- <a href="gg133">LFNG</a>
- <a href="gg132">MFNG</a>
- <a href="gg125">MGAT1</a>
- <a href="gg126">MGAT2</a>
- <a href="gg127">MGAT3</a>
- <a href="gg128">MGAT4A</a>
- <a href="gg129">MGAT4B</a>
- <a href="gg130">MGAT5</a>
- <a href="gg131">MGAT5B</a>
- <a href="gg215">PIGA</a>
- <a href="gg225">PIGB</a>
- <a href="gg214">PIGC</a>
- <a href="gg216">PIGH</a>
- <a href="gg221">PIGM</a>
- <a href="gg217">PIGP</a>
- <a href="gg213">PIGQ</a>
- <a href="gg223">PIGV</a>
- <a href="gg222">PIGX</a>
- <a href="gg218">PIGY</a>
- <a href="gg226">PIGZ</a>
- <a href="gg138">POMT1</a>
- <a href="gg139">POMT2</a>
- <a href="gg134">RFNG</a>
- <a href="gg154">UGCG</a>
-
2.4.2
- <a href="gg169">XYLT1</a>
- <a href="gg170">XYLT2</a>
-
2.4.99
- <a href="gg048">ST3GAL1</a>
- <a href="gg049">ST3GAL2</a>
- <a href="gg050">ST3GAL3</a>
- <a href="gg051">ST3GAL4</a>
- <a href="gg052">ST3GAL5</a>
- <a href="gg053">ST3GAL6</a>
- <a href="gg054">ST6GAL1</a>
- <a href="gg055">ST6GAL2</a>
- <a href="gg056">ST6GALNAC1</a>
- <a href="gg057">ST6GALNAC2</a>
- <a href="gg059">ST6GALNAC4</a>
- <a href="gg062">ST8SIA1</a>
-
2.7.8
- <a href="gg141">DPAGT1</a>
-
2.8.2
- <a href="gg027">CHST1</a>
- <a href="gg036">CHST11</a>
- <a href="gg037">CHST12</a>
- <a href="gg038">CHST13</a>
- <a href="gg028">CHST2</a>
- <a href="gg025">CHST3</a>
- <a href="gg034">CHST4</a>
- <a href="gg029">CHST5</a>
- <a href="gg030">CHST6</a>
- <a href="gg026">CHST7</a>
- <a href="gg045">CHST8</a>
- <a href="gg046">CHST9</a>
- <a href="gg041">GAL3ST1</a>
- <a href="gg042">GAL3ST2</a>
- <a href="gg043">GAL3ST3</a>
- <a href="gg044">GAL3ST4</a>
- <a href="gg024">HS2ST1</a>
- <a href="gg014">HS3ST1</a>
- <a href="gg015">HS3ST2</a>
- <a href="gg016">HS3ST3A1</a>
- <a href="gg017">HS3ST3B1</a>
- <a href="gg018">HS3ST4</a>
- <a href="gg019">HS3ST5</a>
- <a href="gg020">NDST1</a>
- <a href="gg021">NDST2</a>
- <a href="gg022">NDST3</a>
- <a href="gg023">NDST4</a>
-
3.5.1
Open ALL
Close ALL
|



|
About GlycoGene DataBase
Significance of GGDB
Glycogene includes genes associated with glycan synthesis such as glycosyltransferase, sugar nucleotide synthases, sugar-nucleotide transporters, sulfotransferases, etc. At present, over 180 human glycogenes were identified, cloned and characterized. In "Construction of GlycoGene Library Project " (April, 2001 - March, 2004), we collected and compiled the data on such glycogenes as GlycoGene Database (GGDB), which is the first database to store information on substrate specificity. GGDB provides necessary information for the analysis of glycogenes.
Present Status of GGDB
The purpose of GlycoGene Database (GGDB, http://riodb.ibase.aist.go.jp/rcmg/ggdb) is to provide users with easy access to the information on glycogenes via website. In GGDB, the following property information of each glycogene are stored in XML format: gene names(gene symbols), enzyme names, DNA sequences, tissue distribution(gene expression), substrate specificities, homologous genes, EC numbers, and external links to various databases. It graphically shows the information such as substrate specificities, etc.
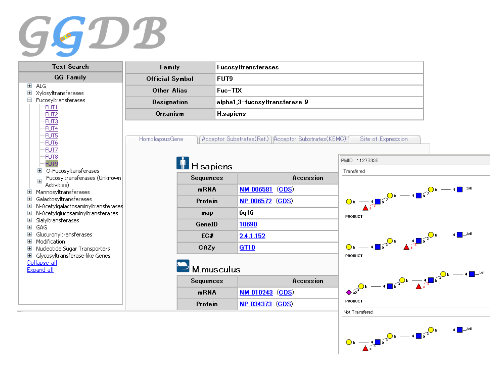
Web Interface of Glycogene Database (GGDB)
Future Perspectives
It is expected that GGDB greatly contributes to improve the efficiency of analysis required for progress in glycobiology. GGDB will be updated to provide more integrated functions for which to display information of enzyme reactions and to link to the mass spectrometry database of glycans. The interface will also be expanded in the future with the addition of other property information, based on users feedbacks. In addition, using data mining technique, we are analyzing the stored data to develop a new tool to predict the substrate specificity for each glycogene.
Acknowledgement
The production of those experimental data is supported by the New Energy and Industrial Technology Development Organization (NEDO).
The construction of this database, the network security and Web
Application Security are supported by Tsukuba Advanced Computing
Center(TACC), AIST.
Disclaimer
Research Center for Medical Glycoscience at AIST does not warrant or assure any legal liability or responsibility for the accuracy, completeness, or usefulness of any information available from this website.
|

 JCGGDB Cross-Search
JCGGDB Structure Search
JCGGDB Cross-Search
JCGGDB Structure Search



