GlycoGene Page
The glycogene page is displayed by selecting Gene Symbol from the list of glycogenes.
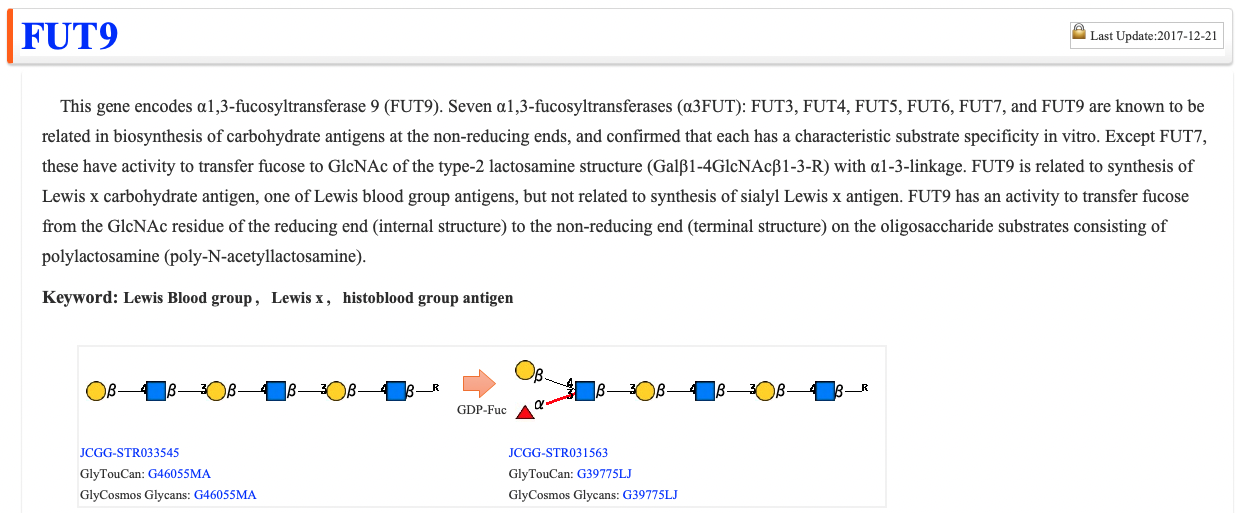
SUMMARY
In this summary, glycogenes are curated by glycoscientists, and enzyme reactions and functions are briefly summarized. The main enzymatic reactions are shown as precursors, substrates (sugar nucleotides) and reaction products as images.
The glycan structures are assigned JCGG-STR as IDs. In addition, links to GlyTouCan and GlyCosmos Glycans are supported.
GGDB uses the HGNC (HUGO Gene Nomenclature Committee) nomenclature for gene names. The common names used so far are stored in Alias. Reference links to external databases are also provided. You can use them to obtain other database information from glycogenes. The following list are items displayed in the summary.
- GGDB Symbol: Symbol names determined by HGNC.
- Alias: Show aliases of glycogene
- HGNC Symbol: Gene symbol determined by HGNC
- historical: Gene symbol used in somewhere past document
- clone name: Clone name, clone id
- related terminology: Named after biological or medical knowledge
- allelic: Symbol for allelic gene
- Alias: Show aliases of glycogene
- Designation: Gene name
- Organism: Organism name
- GeneID: NCBI GeneID and link to NCBI
- HGNC: HGNC ID and link to HGNC
- mRNA: RefSeq ID and link to NCBI
- map: Chromosome location
- Protein: RefSeq ID and link to NCBI
- EC#: EC number and link to IntEnz (Integrated relational Enzyme database)
- CAZy: Family Number and link to CAZy (Carbohydrate-Active enZymes)
- GlyCosmos glycogene: Link to GlyCosmos Glycogene
- OMIM: Link to OMIM
- Disease name: Common disease name
- GDGDB: Link to GDBDB (Glyco-Disease Genes Database)
- Human Protein Atlas: Link to Human Protein Atlas
Orthologous genes
Displays ortholog information of glycogenes in mouse, rat, etc. as NCBI links.
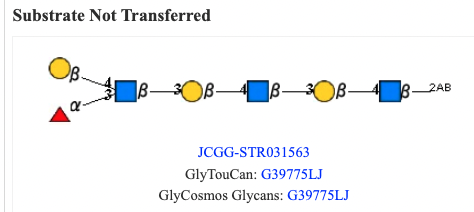
Acceptor Substrates (Reference) & Acceptor Substrates (KEM-C)
Substrate specificity with a glycan structure of the substrate is described as an image together with a glycan structure of the product.
Acceptor information (Reference) is based on the information in the article, and Acceptor information (KEM-C) is based on the database (KEM-C) of substrate specificities of glycosyltransferases.
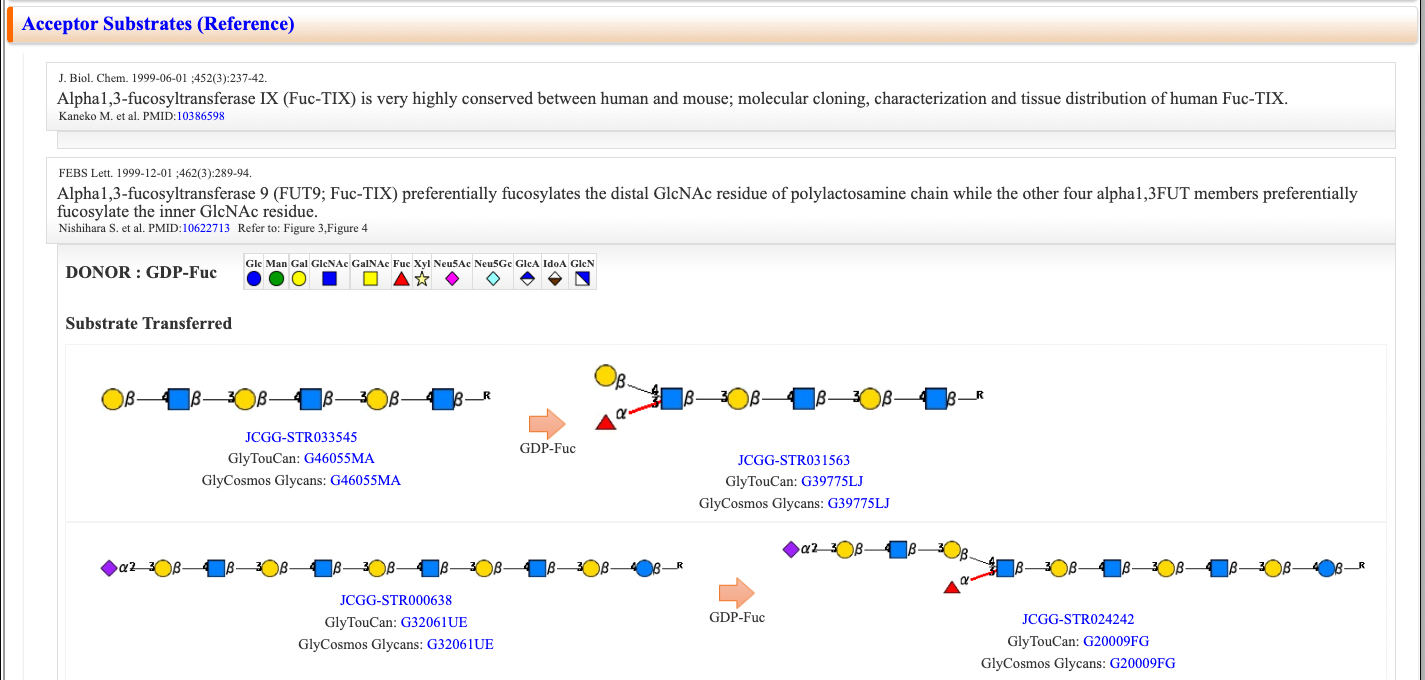
Structures that were not transferred by a enzyme are also indicated.
Expression
RNA-seq data from the Human Protein Atlas are displayed in a bar chart. “Tissue RNA Expression” is a link to the Human Protein Atlas to which this bar chart refers. The version of the RNA-seq data used is also indicated below the bar chart.
Move bar chart left/right & reset
A bar chart can be moved left and right. You can also reset the bar chart by clicking the Home icon.
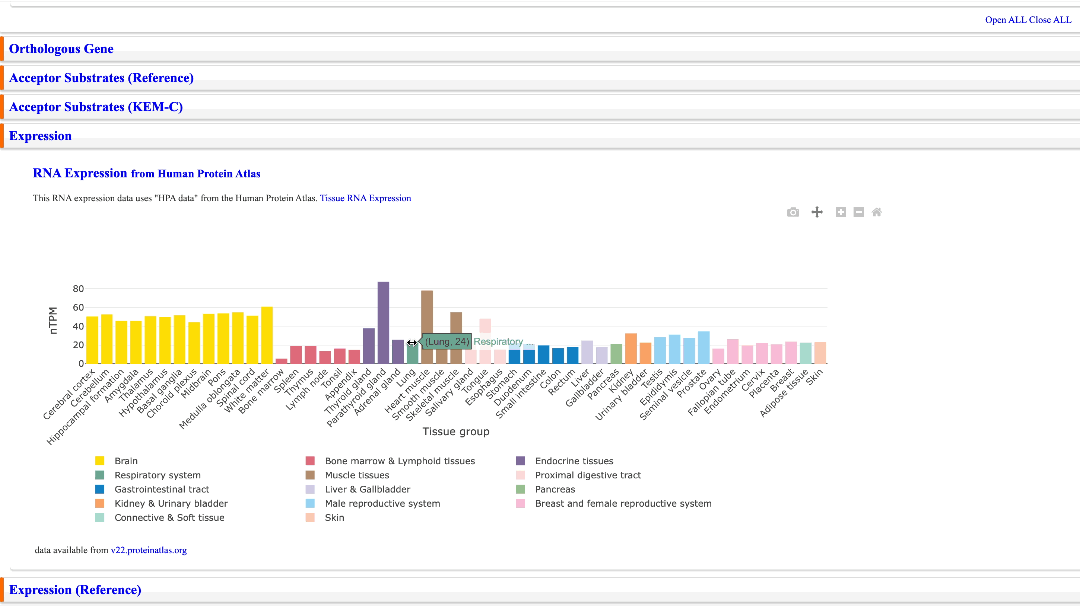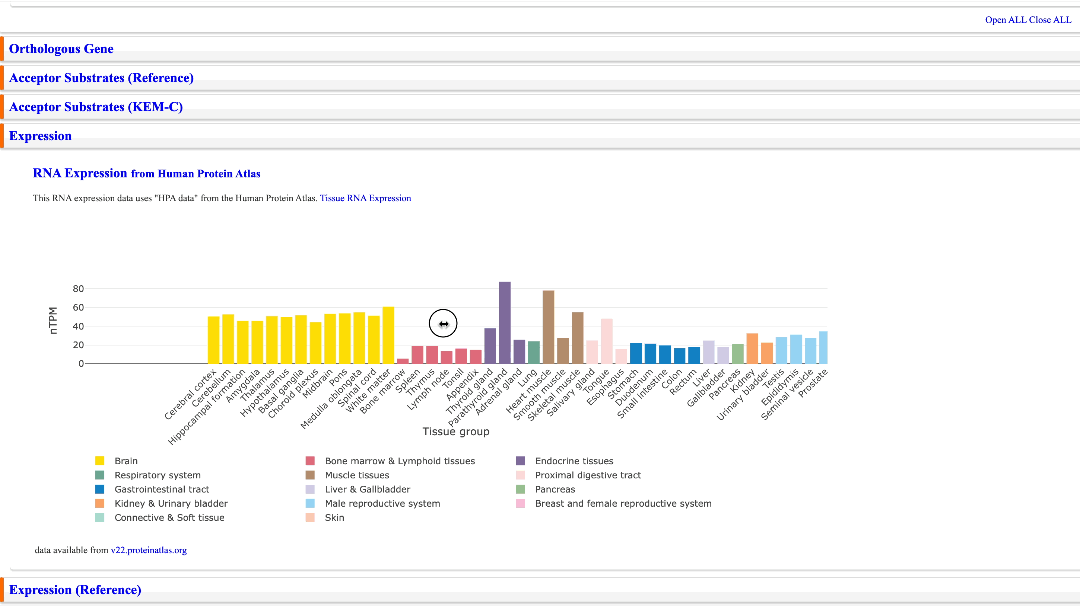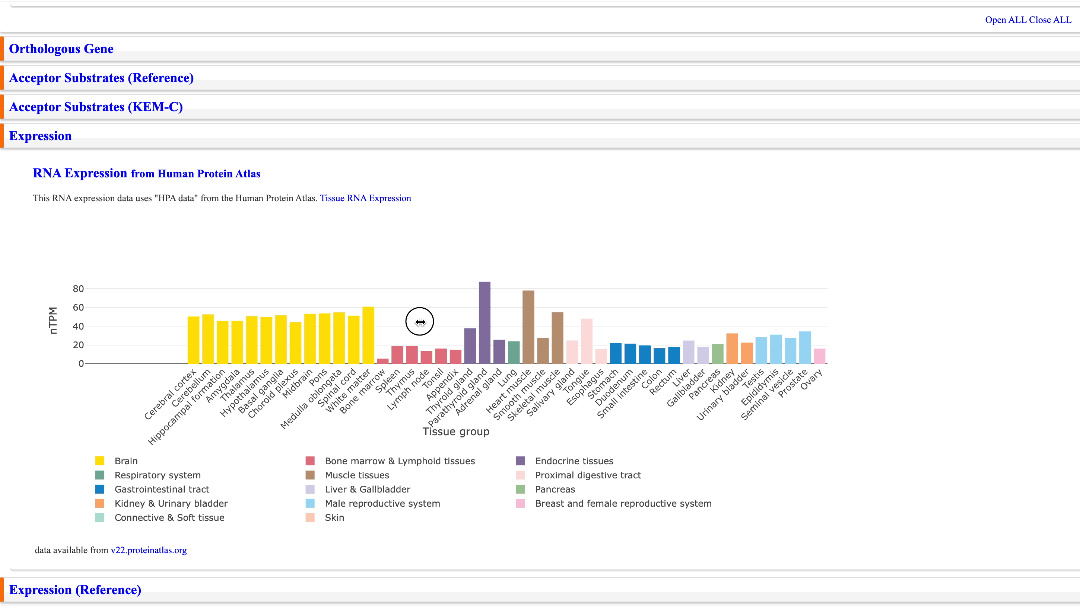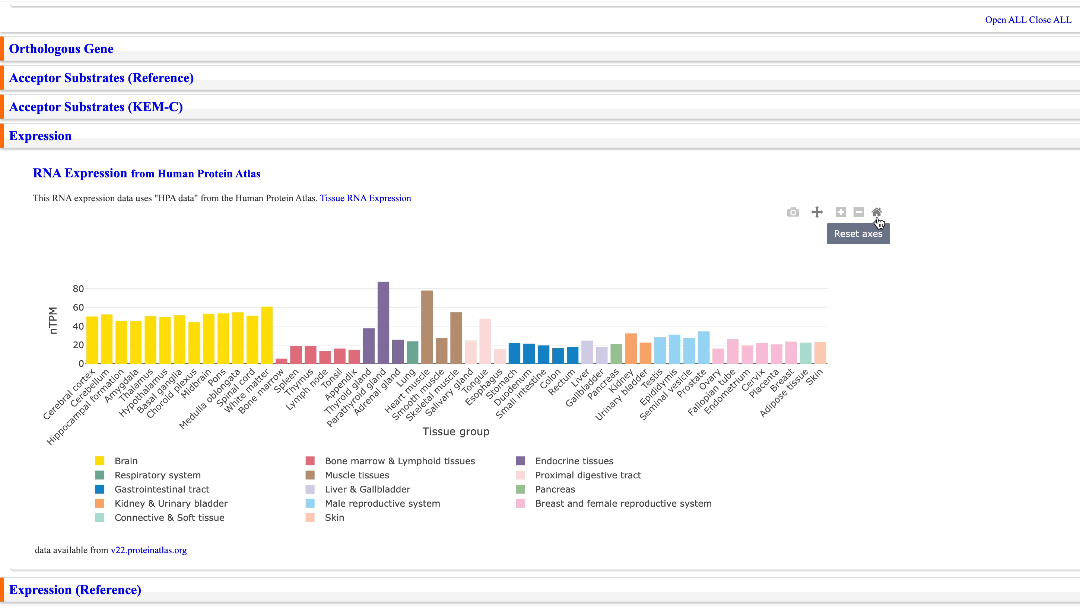
Zoom-in & Zoom-out
Clicking the plus icon zooms the bar chart in. Clicking the minus icon zooms out; you can reset it with the Home icon.
Download bar chart as images
Click on the camera icon to download a bar chart as a png file image. If the camera icon is clicked while the bar chart is being manipulated in some way, the image will be downloaded after the manipulation.
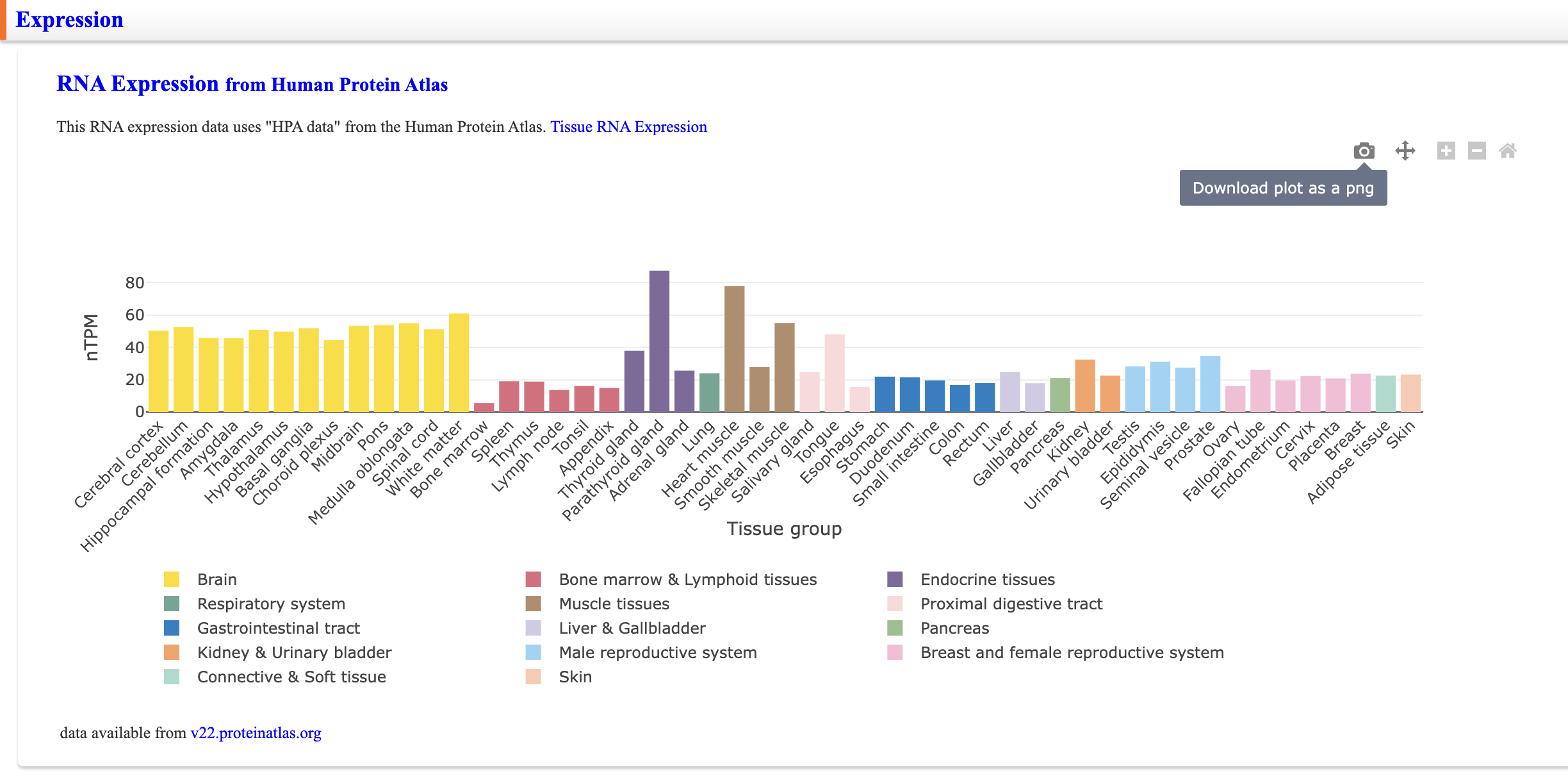
Hide some Tissue Groups
Clicking on any legend below the bar chart will hide the graph of the same color.
Expression (Reference)
Displays articles related to mRNA expression site information.
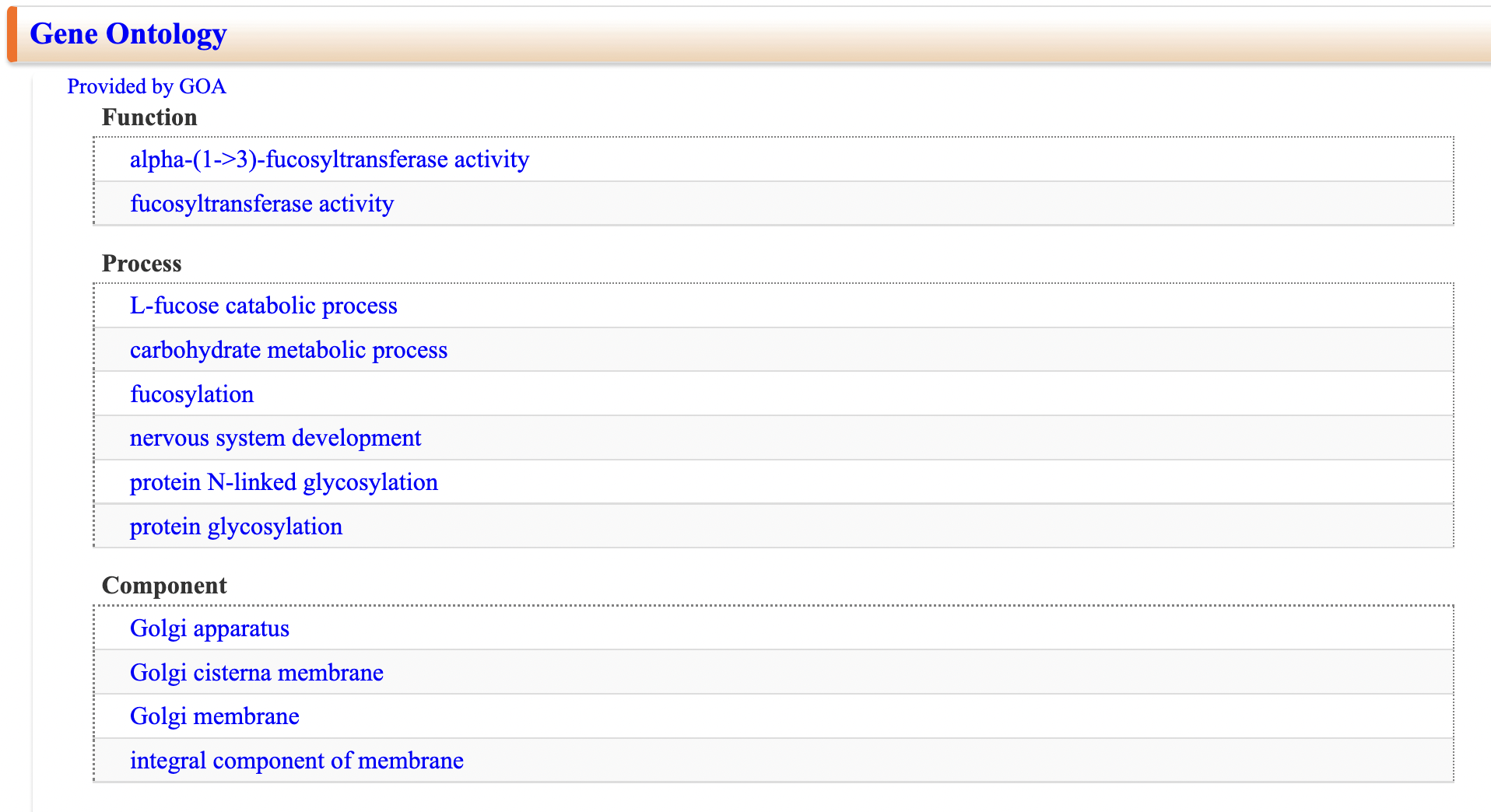
Gene Ontology
Displays Gene Ontology data related to glycogenes. (Provided by Gene Ontology Annotation (GOA) Database)